
|

|
|
|
Projects
Do also take a look at my publications.
ONGOING AND RECENT FUNDED RESEARCH PROJECTS
Principal investigator
- DBT-VINNOVA project 2023-04212 - "A multi-institutional validation study of predicting prostate cancer-specific mortality with AI-based grading"
- Cancerfonden project 22 2353 Pj - "Towards screening for oral cancer: A trustworthy AI-based diagnostic support system for early detection of oral cavity and oropharyngeal cancer"
- AIDA project (VINNOVA through MedTech4Health projects 2017-02447 and 2021-01420) - "What and Why, not just Where - More informative explanations of deep learning-based cancer diagnostics"
- AI4Research project "Explainable AI - What and Why, not just Where: Towards more useful explanations of deep learning-based image classification"
- DBT-VINNOVA project 2020-03611 - "AI-Driven Large-scale Screening for Oral and Oropharyngeal Cancer"
- Centre for Interdisciplinary Mathematics project "Development of Multimodal AI-supported Image Data Analysis Methods for Improved Cancer Diagnostics"
- Swedish Research Council (VR) project 2017-04385 - "Sparse modelling and deep learning for improved Fourier ptychographic microscopy with biomedical applications"
- AIDA project (VINNOVA through MedTech4Health project 2017-02447) - "Trustworthy AI-based decision support in cancer diagnostics"
Additionally supported by Swedish National Infrastructure for Computing (SNIC) through Projects SNIC 2021/7-58, SNIC 2021/7-128 - AIDA project (VINNOVA through MedTech4Health project 2017-02447) - "Image- and AI-based cytological cancer screening"
- VINNOVA Innovation project 2015-01667 - "Predictive modelling of real time video of outdoor scenes captured with a moving handheld camera"
- Swedish Research Links project 2015-05878 - "Image analysis for reliable and cost effective cancer detection"
Participant
- SOF (Styrgruppen för odontologisk forskning) project grant 2024-2026, FoUI-991579 - "AI, biomarkers, and precision medicine for prevention, diagnostics, and treatment of oral cancer"; PI: Carina Kruger Weiner
- AIDA project (VINNOVA through MedTech4Health project 2017-02447 and 2021-01420) - "AI-driven analysis and knowledge transfer between multiplex and brightfield microscopy"; PI: Nataša Sladoje
- Region Stockholms Innovationsfond, FoUI-985869 - "Riktad screening för att minska insjuknandet i munhålecancer"; PI: Ronak Sandy
- Swedish Research Council (VR) project 2022-03580 - "Interpretable AI-based multispectral 3D-analysis of cell interrelations in cancer microenvironment for patient-specific therapy"; PI: Nataša Sladoje
- Cancerfonden project 22 2357 Pj - "From cancer biology to patient stratification: Interpretable AI-based multispectral 3D-analysis of cell interrelations in the cancer microenvironment"; PI: Nataša Sladoje
- eSSENCE-SciLifeLab project - "OncoSpace - multispectral 3D-analysis of immune cell interaction in the lung cancer microenvironment for patient-specific cancer therapy"; PI: Patrick Micke
- Region Stockholms Innovationsfond, FoUI-973302 - "Riktad screening för att minska insjuknandet i munhålecancer"; PI: Carina Kruger Weiner
- FTV Stockholm AB, Prioritized research project "AI-baserat beslutsstödsystem för tidig upptäckt av muncancer"; PI: J.-M. Hirsch
- AIDA project (VINNOVA through MedTech4Health project 2017-02447) - "Multimodal imaging and information fusion for confident image-based cancer diagnostics"; PI: Nataša Sladoje
- Centre for Interdisciplinary Mathematics project "Explainable deep learning methods for human-human and human-robot interaction"; PI: Ginevra Castellano
- WASP, Wallenberg AI, Autonomous Systems and Software Program, project "Robust joint learning and utilization of geometric equivariances for biomedical image analysis", within the WASP-AI/MATH research program; PI: Nataša Sladoje
- CA COST Action CA17121 - "Correlated Multimodal Imaging in Life Sciences (COMULIS)"; MC Substitute
- CA COST Action CA15124 - "A new Network of European BioImage Analysts to advance life science imaging (NEUBIAS)"; MC Member
- VINNOVA MedTech4Health project 2016-02329 - "En smart och lättanvänd plattform för att underlätta ultrastrukturella patologiska diagnoser"
- "Advanced Techniques of Cryptology, Image Processing and Computational Topology for Information Security"; Grant ON 174008 of the Ministry of Science and Technological development of the Republic of Serbia.
- "Development of new information and communication technologies, based on advanced mathematical methods, with applications in medicine, telecommunications, power systems, protection of national heritage and education"; Grant III 44006 of the Ministry of Science and Technological development of the Republic of Serbia.
- "Collaborative development of methods for robust and precise image analysis for cost effective and reliable detection of cervical cancer." Grant nr. 2014-4231, Swedish Research Council, within Swedish Research Links program.
Past and current projects at CBA
- Fuzzy shape analysis in 2D and 3D
Ingela Nyström, Joakim Lindblad, Gunilla Borgefors
Funding: SLU S Faculty, UU TN Faculty
Period: 0109-
Partners: Nataša Sladoje (Matic), Faculty of Engineering, University of Novi Sad, Serbia;
Abstract: The advantages of representing objects in images as fuzzy spatial sets are numerous and have lead to increased interest for fuzzy approaches in image analysis. Fuzziness is an intrinsic property of images and a natural outcome of most imaging devices. Preservation of fuzziness implies preservation of important information about objects and images. Our previous results within this project show that an improved precision of a shape description can be achieved if the description is based on a fuzzy, instead of a crisp shape representation, where the fuzzy membership of a point reflects the level to which that point belongs to the object.During 2006, a manuscript on the representation and reconstruction of fuzzy disks by moments, containing derivation of theoretical error bounds for the accuracy of the estimation of moments of a continuous fuzzy disk from the moments of its digitization, as well as showing that, for a certain class of membership functions, there exists a one-to-one correspondence between the set of fuzzy disks and the set of their generalized moment representations, was accepted for publication in the Fuzzy Sets and Systems journal.
- Defuzzification of fuzzy segmented objects by feature invariance
Ingela Nyström, Joakim Lindblad, Stina Svensson
Funding: SLU S Faculty, UU TN Faculty
Period: 0301-
Partners: Nataša Sladoje (Matic) and Tibor Lukic, Faculty of Engineering, University of Novi Sad, Serbia
Abstract: This project concerns the development of a method for feature based defuzzification of spatial fuzzy sets. The developed method generates crisp shapes from fuzzy shapes by finding a crisp shape at a minimal distance to the fuzzy shape. We define the distance between two fuzzy sets as a distance between their feature-based representations in a chosen feature space. We have found it appropriate for defuzzification to incorporate both local and global features of the two sets. We have studied the use of membership values, gradient, area, perimeter, and centre of gravity in the distance measure. Several existing distance measures can be used to define the distance measure in the feature space. We have so far focused the research on Minkowski type distances measure.
The defuzzification method was further developed during 2006. A method for generating the crisp discrete representation of a fuzzy set at an increased spatial resolution, compared to the resolution of the fuzzy set, was developed and presented at the IWCIA conference in June 2006. Additional refinement of the method was achieved by the use of a scale space approach, providing preservation of feature values over a range of scales in the defuzzification process. Initial results from such an approach, where area at a range of scales was used in the feature distance, were presented at the DGCI conference in October 2006. That presentation also included a practical implementation of the method for 3D data. An example of defuzzification of a 3D data set at increased resolution is presented in Figure 4.
Ongoing research regarding improvements of the optimization part of the method, as well as practical application of the defuzzification method to Cryo-ET data of proteins. was also undertaken during 2006.Figure 4: Defuzzification of a bone region from a 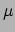 CT volume image.
Slice through the image volume (a).
Slice through a fuzzy segmentation of the bone region in the image volume (b).
Slice through a defuzzification, using meso-scale volume features of the fuzzy segmented object (c).
3D rendering of the
CT volume image.
Slice through the image volume (a).
Slice through a fuzzy segmentation of the bone region in the image volume (b).
Slice through a defuzzification, using meso-scale volume features of the fuzzy segmented object (c).
3D rendering of the 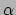 -cut at smallest feature distance to the fuzzy object (d).
3D rendering of a high resolution defuzzification of the fuzzy segmented object (e).
The values
-cut at smallest feature distance to the fuzzy object (d).
3D rendering of a high resolution defuzzification of the fuzzy segmented object (e).
The values 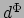 below images (d) and (e) indicate the feature distance of the corresponding object
to the original fuzzy image.
below images (d) and (e) indicate the feature distance of the corresponding object
to the original fuzzy image.
- New objective quantitative analysis techniques for quantification of tissue regeneration around medical devices
Gunilla Borgefors, Joakim Lindblad, Hamid Sarve
Funding: SLU, S Faculty, Swedish Research Council
Period: 0503-
Partner: Carina Johansson, Dept. of Clinical Medicine, Örebro University
Abstract: In order to evaluate how tissue reacts on implants, the interface between the implant and the tissue must be studied. Today, this procedure is done manually in a microscope.
The aim of this project is to develop automatized image analysis methods for analyzing images of the junction of tissue and implant. This method shall make the procedure more effective as well as giving an objective estimation.
The analysis involves segmentation of the images in different tissue-types and measurement of some relevant measures such as length, area and volume.
Before the analysis, methods that shall remove artifacts be applied. Differences in graylevels, color and possibly texture features will be used for the recognition. Known methods will be used to present the result. The interpretation of the values however, will not be done by the postgraduate student.
This project will result in a number of publications (at conferences and in technical and medical journals) about the new methods used as well as the resulting measurement. - Analysis of skeletal muscle fibers in 3D images
Patrick Karlsson, Ewert Bengtsson, Joakim Lindblad, Gunilla Borgefors
Period: 0603-
Partners: Anna-Stina Höglund, Jingxia Liu, Lars Larsson, Dept. of Neuroscience, UU
Abstract: The need for understanding of the three dimensional (3D) spatial arrangement of myonuclei in skeletal muscle fibers is great. A highly detailed 3D spatial description of the organization of myonuclei in healthy and diseased human muscle cells enables detailed understanding of the underlying mechanisms of muscle wasting associated with, e.g., neuromuscular disorders, and aging. The current poor understanding of the spatial arrangement of myonuclei is to be remedied by an interdisciplinary collaboration between the CBA and the Muscle Research Group (MRG) at UU. This project develops and evaluates methods for modeling and quantitative analysis of 3D distributions of myonuclei by utilizing the proficiency of modern confocal microscopic techniques to create true 3D volume images. Advanced computerized modeling of the elongated generalized cylinder structure of the imaged muscle cells is paramount in investigating the myonuclear domain, i.e., each finite volume in which a myonucleus control the gene products. The complex topological conditions put on 3D models and measurement methods, compared to 2D equivalents, are a challenge in developing appropriate image analysis tools and algorithms. The project was presented with a poster presentation at "Medicinteknikdagarna 2006'' in Uppsala.Figure 5: A: A 2D-slice of a skeletal muscle fiber from rat. The red signal represents the fiber sarcomere, and the blue represents the nuclei. B: A maximum intensity projection of the nuclei perpendicular to the imaging plane. C: Iso-surface of the fiber signal. D: 3D-view of the nuclei signals with estimated centroid position based on local intensity maximums. E: Retrieved nuclei centroids, based on D. 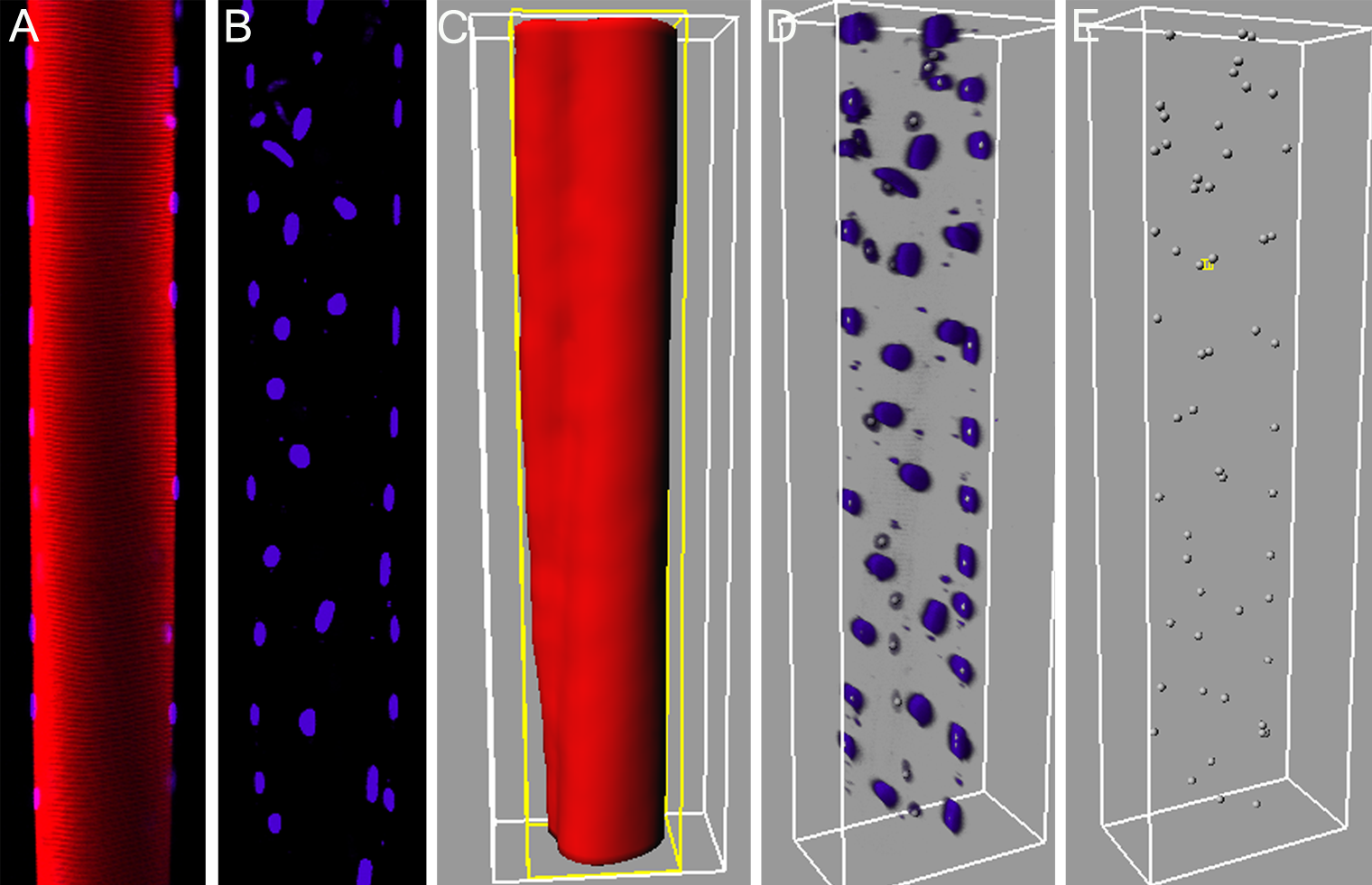
- Image analysis of the internal structure of paper and cellulose based composite materials in 3D images
Maria Axelsson, Filip Malmberg, Stina Svensson, Joakim Lindblad, Gunilla Borgefors
Funding: SLU S Faculty
Period: 0406-
Partners: Norwegian Pulp and Paper Research Institute (PFI), Trondheim, Norway; STFI-Packforsk, Stockholm; KTH Solid Mechanics, Stockholm; StoraEnso, Falun
Abstract: The internal structure of paper is important to study since many material properties correspond directly to the properties of single fibres and their interaction in the fibre network. How single fibres in paper bond and how this effects paper quality is not fully understood since most structure analysis of paper has been performed in cross sectional two dimensional (2D) images and paper is a complex three dimensional (3D) structure. Another application for wood-fibres that has recently gained a lot of interest is wood polymer composite materials. The properties of these materials do not only depend on the structure of the fibre network, but also on interaction between the fibres and the polymer matrix surrounding the fibres. Advances in imaging technology has made it possible to acquire 3D images of paper and wood polymer composite materials. In this project, image analysis methods for characterising the 3D material structure in such images are developed. The detailed knowledge of the material structure attainable with these methods is useful for improving material properties and for developing new materials.
An example slice from a binarised volume and a surface rendering of a sample of a composite material image with X-ray microtomography are shown in Figure 15.
The project objective is to achieve a complete segmentation of individual fibres and pores in volume images of the material. Given such a segmentation, any measurement of the internal structure is available. Measurements on individual fibres and the structural arrangement of fibres can then be related to macroscopical material properties. Other methods for measuring properties of the material, that do not require a complete segmentation of the samples, are also investigated.
In this project, different volume images of paper and composite materials are available for the studies. This includes one volume created from a series of 2D scanning electron microscopy (SEM) images at StoraEnso in Falun and X-ray microtomography volume images of paper and composite samples imaged at the European Radiation Synchrotron Facility (ESRF) in Grenoble, France. Furthermore, methods for creating other sample volume images are investigated.
During 2006, the project has resulted in a number of publications. A method for reducing ring artifacts in the volume images using orientation estimates and normalised convolution in the polar domain was presented at the DAGM conference in September 2006 and published in the Lecture Notes in Computer Science Series. See Figure 16 for an illustration of the method. At the Progress in Paper Physics Seminar three extended abstracts were presented. One abstract on 3D structure characterisation of newsprints, one on stress transfer and failure in pulp fibre reinforced composite materials and one on damage mechanisms in paper.Figure 15: A slice from a binarised volume image of a composite material (left). A surface rendering of a sample of a composite material (right). - Image analysis for quantitative estimation of seed vitality
Joakim Lindblad, Gunilla Borgefors
Funding: The Swedish Farmers' Foundation for Agricultural Research (SLF)
Period: 0509-0603
Partners: AnalyCen Nordic AB, Lidköping; SeedGard AB (prev. Acanova), Uppsala; Anders Larsolle, Dept. of Biometry and Engineering, SLU, Uppsala
Abstract: ThermoSeed cereal seed treatment is a new method for thermal seed treatment developed by SeedGard AB (prev. Acanova). The method makes it possible to produce seed free from seed-borne pathogens without using chemical seed dressing. By exposing seeds for precisely conditioned hot humid air, pathogens are rendered harmless without affecting seed germinability.
It is of interest to facilitate objective and accurate monitoring of how different treatments and different types of stress affects the vitality of seeds. During the autumn of 2005 computer software for automatic segmentation and separation of individual plants as well as to measure relevant parameters, such as area and length, of the plants were developed. In early 2006, the software was delivered to SeedGard AB, where further testing and tuning of parameters followed. - Algorithms for segmentation of fluorescence labelled cells
Carolina Wählby (née Linnman), Joakim Lindblad, Mikael Vondrus, Ewert Bengtsson
Funding: Amersham Pharmacia Biotech, UU TN-faculty
Period: 9902-
Partners: Lennart Björkesten, Amersham Pharmacia Biotech, Uppsala; Torsten Jarkrans, Dept. of Information Technology, UU
Abstract: The interaction with and effect of potential drugs on living cells can be observed by fluorescence microscopy. Automated methods for feature extraction from fluorescence microscopy images of cells can be used as a tool in the drug discovery process. The cell nucleus has a well-defined shape and is relatively easy to detect. The cytoplasm is however more complex. The first goal of this project was to develop a fully automatic method for cytoplasm segmentation. The present algorithm, inspired by literature and previous experience, consists of an image pre-processing step, a general segmentation and merging step followed by a quality measure and a splitting step. By training the algorithm on one image, it is made fully automatic for subsequent images created under similar conditions. This method was presented at an internal Amersham Pharmacia Biotech R&D conference in Uppsala in late 1999.
During 2000 the algorithms were improved through a more elaborated shape analysis and a more consistent feature extraction and quality evaluation step. The results were documented in a journal paper that was prepared for submission in early 2001. - Estimation of intensity non-uniformities in 2D and 3D microscope images
Joakim Lindblad
Funding: UU TN-faculty
Period: 0001-
Abstract: Intensity non-uniformity (INU), also commonly referred to as shading artifacts, is a common problem in computerized image analysis. Although the INU has limited impact on visual interpretation, it may have a negative effect on image segmentation as well as the interpretation of image intensity values. There exists a range of methods to perform correction of intensity non-uniformities. This project is aimed both at evaluating which methods performs best and under what conditions, and towards finding possible improvements to the existing methods. Although trying to stay general the project is biased toward the applicability to microscope images, mainly of fluorescence stained cells. Under the scope of this project a method to reduce the parameter space of a B-spline based iterative shading estimation algorithm has been suggested and evaluated. - Computer assisted cervical cytology
Ewert Bengtsson, Bo Nordin, Mikael Vondrus, Joakim Lindblad
Funding: AccuMed International, Inc., Chicago, USA
Period: 9407-
Abstract: The computer assisted or automated analysis of cell specimens from the uterine cervix, so called PAP-smears, obtained during gynecological health control is one of the long standing problems in digital image analysis. One of the groups that formed the CBA was active in this field 1973-1987. Since the Summer of 1994 we have had a cooperation project with the Chicago based company AccuMed International, Inc. A number of student projects and some investigations by the senior researchers at CBA have been carried out within this cooperation.
During the last few years the AccuMed company has changed its focus to become a more general supplier of technology for computerized microscopy for various applications. We have discussed projects in which CBA could become involved in renewed cooperation in studying basic methodological problems in this field. Most recently Ewert Bengtsson visited Chicago for two days in March 2000 but these discussions have not yet led to new research activity in Uppsala. - Methods to perform automatic selection of optimal or near optimal threshold values
Joakim Lindblad
Funding: UU TN-faculty
Period: 0001-
Abstract: The simplest and by far most popular method of separating data into different classes is by thresholding. Although not very elaborate and often too abrupt, it is robust and performs well in many different situations. The problem of selecting thresholds is very general and applicable in a vast range of problems. Still there exist no satisfying general solution to the threshold selection problem. This project addresses the task of finding robust all purpose methods for selecting good thresholds in a general distribution. Inspired by statistics, the use of Kernel Density Estimates (KDE) to threshold distributions at locations of high second derivative has been investigated and shown to be fruitful. The difficult problem still to be solved relates to optimal selection of the width of the kernel function for the estimate. The KDE approach was presented at the SSAB conference in March 2000 in Halmstad.
|