Current research projects (2018), Robin Strand
See also projects from the CBA Annual Report.
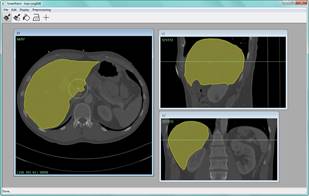 Interactive
image processing
Interactive
image processing
Digital imaging
technique such as whole-slide scanning, computed tomography (CT) and magnetic
resonance imaging (MRI ) are now routinely used in
medicine. This has led to an ever increasing flow of high-resolution image data
that needs to be qualitatively and quantitatively analyzed. We develop powerful
new methods for interactive image processing (including quantification,
segmentation and registration) in collaboration with medical experts.
Key
references:
· Filip Malmberg, Richard Nordenskjöld, Robin Strand, and Joel Kullberg SmartPaint - A Tool for Interactive Segmentation of Medical
Volume Images Computer Methods in Biomechanics and Biomedical Engineering:
Imaging & Visualization, Volume 5, Issue 1, Pages 36-44, 2017
· Andreas Kårsnäs, Robin Strand, Johan
Dore, Thomas Ebstrup, Michael Lippert, Kim Bjerrum A
histopathological tool for quantification of biomarkers with subcellular
Resolution Computer Methods in Biomechanics and Biomedical Engineering: Imaging
& Visualization, Volume 3, Issue 1, 2015, Pages 25-46
· Andreas Kårsnäs and Robin Strand
Multimodal histological image registration using locally rigid transforms In
proceedings of Interactive Medical Image Computation (IMIC), MICCAI 2015
workshop, Munich
· Filip Malmberg, Robin Strand, Joel
Kullberg Interactive Deformation of Volume Images for Image Registration In
proceedings of Interactive Medical Image Computation (IMIC), MICCAI 2015
workshop, Munich
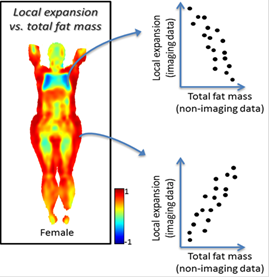 Large
scale whole body image processing
Large
scale whole body image processing
PhD
students: Simon
Ekström, Therese Sjöholm, Eva Breznik, Martino Pilia
Imiomics
is an image processing concept that consists of a set of methods, including
image registration, that allow statistical and holistic analysis of whole-body
image data and non-imaging data. Imiomics enables
creation of a Human Imaging Atlas, a statistical representation of intra-group
distributions of image features. Imiomics analyses
are holistic for three reasons: 1) the whole body is analyzed,
2) all collected image data is used in the analysis, and 3) it allows
integration of all other collected non-imaging patient information in the
analysis. The image registration method used utilizes quantitative whole-body
water-fat MRI data, a pre-segmentation of bone from these images, and tissue
specific constraints in the registration process.1 Imiomics
supports inclusion of other image data, such as DWI or PET, in the analysis.
Potential applications of Imiomics include 1) to
compare whole-body image feature between groups of for example sick and healthy
subjects, 2) to follow changes in whole-body images in a subject over time,
e.g. after intervention, 3) to assist attenuation correction in PET-MR where
separation of bone and air is challenging, 4) to allow calculation of whole
body images of point-by-point or tissue-by-tissue statistical interaction
between imaging and non-imaging features, e.g. a correlation map between
insulin levels and morphology like regional adipose tissue or muscle tissue
volumes.
Key
references:
· Robin Strand, Filip Malmberg, Lars
Johansson, Lars Lind, Magnus Sundbom, Håkan Ahlström, Joel Kullberg A Concept
for Holistic Whole Body MRI Data Analysis, Imiomics
PLOS ONE 12(2): e0169966, 2017
· Joel Kullberg, Håkan Ahlström, Robin
Strand, WHOLE BODY IMAGE REGISTRATION METHOD AND METHOD FOR ANALYZING IMAGES
THEREOF Patent application WO 2016/072926
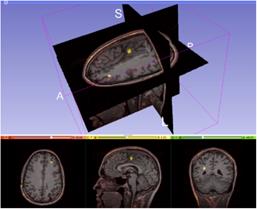 Subtle
change detection
Subtle
change detection
Post
Doc: Ashis Kumar
Dhara
In this
project, semi-automatic tools for fast and precise magnetic resonance volume
image processing and analysis will be developed for change detection in
traumatic brain injury, neurodegenerative diseases including intracranial
aneurysms and brain tumors.
Subtle
change detection and quantification is a challenging problem due to limited
resolution, partial volume effects, noise, artefacts, etc. Detection of small
volumetric changes with high confidence is very important for diagnosis and for
selection of, and to follow up the effect of, treatment.
Automatic
or semi-automatic image processing methods are needed due to the difficulty to
detect subtle volume changes by visual inspection.
Radiotherapy using integrated MR and Linac
PhD
student: Samuel
Fransson
A combined
magnetic resonance scanner and radiotherapy treatment unit will be installed at
Akademiska hospital in Uppsala during 2018. This will
be one of the first installations in the world of this next generation
radiotherapy treatment unit, developed by the Swedish company Elekta.
We develop
software to support treatment of small treatment volumes moving in an irregular
way. Examples of this kind of volume are the prostate gland, individual lymph
nodes and radioresistent subvolumes
within larger tumors.
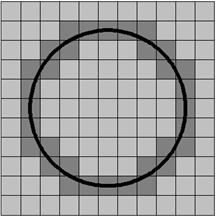 Digital
geometry
Digital
geometry
In a wide sense,
digital topology and geometry refers to the use of topologic and geometric
properties and features for images defined in digital grids. Our research in
this area focuses on methods where the theory and algorithms use the principles
of digital path connectivity, path propagation, and neighborhood analysis,
often by pixel adjacency graph representations. The methods are often developed
for medical image processing applications.
Key
references:
· Punam K. Saha, Robin Strand, Gunilla
Borgefors Digital Topology and Geometry in Medical Imaging: A survey IEEE
Transactions on Medical Imaging, Volume 34, Issue 9, 2015, Pages 1940-1964
· Robin Strand, Krzysztof C.
Ciesielski, Filip Malmberg, Punam K. Saha The Minimum Barrier Distance Computer
Vision and Image Understanding, Volume 117, Issue 4, 2013, Pages 429-437
· Robin Strand, Benedek Nagy and
Gunilla Borgefors Digital Distance Functions on Three-Dimensional Grids Theoretical
Computer Science, Volume 412, Issue 15, 2011, Pages 1350-1363
· Robin Strand Distance Functions and
Image Processing on Point-Lattices With Focus on the 3D Face- and Body-centered
Cubic Grids ACTA UNIVERSITATIS UPSALIENSIS Uppsala Dissertations from the
Faculty of Science and Technology, ISSN 1104-2516; 79 ISBN 978-91-554-7303-7
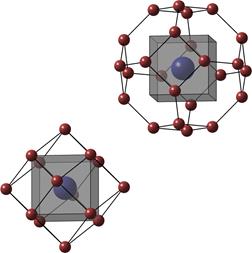 Alternative
sampling grids
Alternative
sampling grids
PhD
students: Elisabeth
Linnér (PhD 2015), Teo Asplund
When
using optimal sampling grids, such as the 2D hexagonal or 3D fcc or bcc grids, fewer samples
can be used to represent images with changing the reconstruction/representation
quality. We develop methods for image acquisition, processing and visualization
on high-dimensional, non-standard grids. Mathematical Morphology methods on irregular
grids are also developed.
Key
references:
· Robin Strand Distance Functions and
Image Processing on Point-Lattices With Focus on the 3D Face- and Body-centered
Cubic Grids ACTA UNIVERSITATIS UPSALIENSIS Uppsala Dissertations from the Faculty
of Science and Technology, ISSN 1104-2516; 79 ISBN 978-91-554-7303-7
· Teo Asplund, Cris L. Luengo
Hendriks, Matthew John Thurley, Robin Strand Mathematical Morphology on
Irregularly Sampled Data in One Dimension Mathematical Morphology-Theory and
Applications, Volume 2, Issue 1, Pages 1-24, 2017. DOI: https://doi.org/10.1515/mathm-2017-0001
· Céline Fouard,
Robin Strand and Gunilla Borgefors Weighted Distance Transforms Generalized to
Modules and their Computation on Point Lattices Pattern Recognition, Volume 40,
Issue 9, September 2007, pages 2453-2474
· Elisabeth Schold Linnér, Max Morén,
Karl-Oskar Smed, Johan Nysjö, Robin Strand LatticeLibrary and BccFccRaycaster:
Software for processing and viewing 3D data on optimal sampling lattices, SoftwareX, Volume 5, Pages 16–24 2016
Current
collaboration partners:
Håkan Ahlström,
Faculty of Medicine, Uppsala University
Joel
Kullberg, Faculty of Medicine, Uppsala University
Punam Saha,
University of Iowa, US
Krzysztof
C. Ciesielski, West Virginia University and University of Pennsylvania
Tufve
Nyholm, Medical Faculty, Uppsala University
Benedek
Nagy, Eastern Mediterranean University, Turkey
Filip Malmberg, Uppsala University
Johan
Wikström, Faculty of Medicine, Uppsala University
Elna-Marie
Larsson, Faculty of Medicine, Uppsala University